H5AD DATASET FILE FORMAT
WHAT IS IT?
H5AD is a file format used primarily for storing large-scale single-cell data, in a hierarchical manner. It is based on the HDF5 format (Hierarchical Data Format version 5) with the AnnData structure in Python, used mainly in bioinformatics and computational biology.
WHAT IS ITS STRUCTURE?
H5AD files can store complex data structures, including arrays, key-value pairs, tabular data, and metadata in a single file. The format also supports data compression.
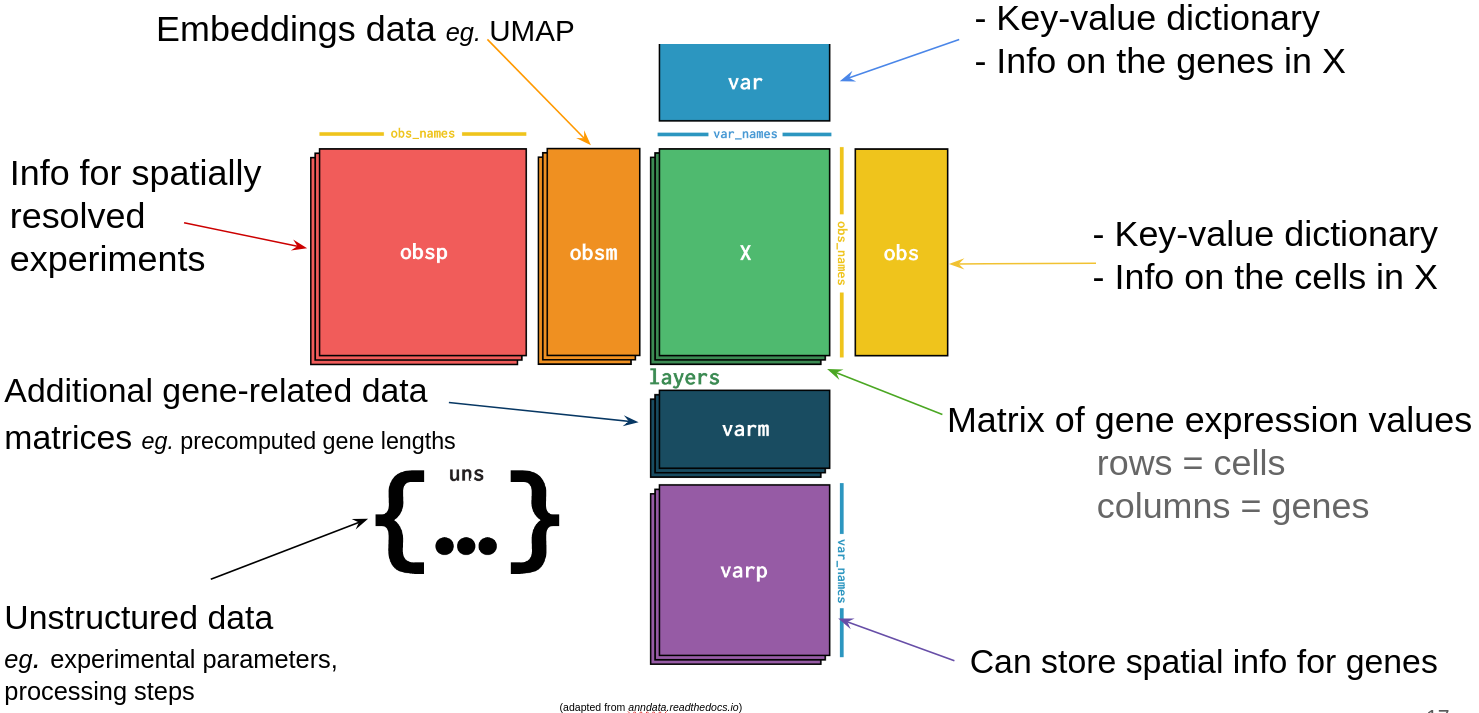
Components (let’s call them tables that hold annotations):
Note: var and obs can also be Pandas DataFrame
HOW TO PREVIEW IT?
Code 1:
```
import scanpy as sc
adata = sc.read_h5ad("your_file.h5ad")
print(adata) # summary
print(adata.X.shape) # matrix dims
print(adata.obs.head()) # cell metadata
```
Code 2:
```
from anndata import read_h5ad
adata = read_h5ad("your_file.h5ad")
print(adata) # summary
print(adata.var.head()) # gene metadata
```
Code 3:
```
import h5py
f = h5py.File("your_file.h5ad","r")
print(list(f.keys())) # top‐level groups: X, obs, var, uns, …
print(f["X"].shape) # expression mat dims
print(list(f["obs"].keys())) # obs columns
f.close()
```
```
library(hdf5r)
f <- H5File$new("your_file.h5ad", mode = "r")
names(f) # top‐level groups: X, obs, var, uns, …
f[["X"]]$dims # expression mat dims
names(f[["obs"]]) # obs columns
f$close_all()
```
You can also use sceasy, Seurat, or zellkonverter
HOW TO CREATE IT?
You can build the file from your cell and gene data by using Python libraries such as anndata and adhering to the format structure outlined above.
Additionally, you can convert between formats, eg. from H5 to H5AD, by using conversion functions provided by libraries specific to H5AD (such as the ones used to preview it) or to the file format you are currently using.
Feel free to reach out with any questions.